Журнал "Гастроэнтерология" Том 58, №4, 2024
Вернуться к номеру
Посттрансляційні модифікації гістонів, що пов’язані з розвитком метаболічно асоційованої жирової хвороби печінки. Частина 2. Метилювання гістонів
Авторы: Абатуров О.Є., Нікуліна А.О.
Дніпровський державний медичний університет, м. Дніпро, Україна
Рубрики: Гастроэнтерология
Разделы: Справочник специалиста
Версия для печати
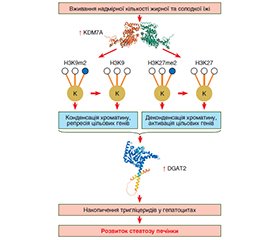
На основі аналізу літературних джерел баз даних Pubmed, MedLine, The Cochrane Library, EMBASE автори статті висвітлюють істотну роль епігенетичних модуляцій у патогенезі метаболічно-асоційованої жирової хвороби печінки. Наведено загальні положення стосовно посттрансляційних модифікацій гістонів, які пов’язані з розвитком метаболічно асоційованої жирової хвороби печінки, а саме особливостей їх метилювання, посттрансляційних модифікацій метилювання лізинових залишків (сайтів H3K4, H3K27, H3K36, H3K79, H4K20, H3K23, H3K63 та H4K12), аргінінових залишків (сайтів H2AR3, H4R3, H3R2, H3R8 і H3R26). Метилювання та деметилювання гістонів, опосередковуючи експресію ключових генів, що беруть участь в обміні вуглеводів та ліпідів, визначають як виникнення, так і розвиток стеатозу, запалення та фіброзу печінки у хворих на метаболічно асоційовану жирову хворобу печінки. Метильовані маркери H3K9, H3K27, H4K20 гістонів пов’язані з упакованим гетерохроматином і репресією транскрипції генів, тоді як метильовані маркери H3K4, H3K36, H3K79 гістонів — з активацією транскрипції генів. Метилювання лізинових або аргінінових залишків гістонів здійснюється метилтрансферазами, які використовують S-аденозилметіонін як донорську молекулу. Метильований сайт аргінінового залишку може бути представлений монометильованим, асиметрично диметильованим або симетрично диметильованим маркером, тоді як видалення метилювання лізинового маркера опосередковується деметилазами. Автори наводять дані, що при метаболічно асоційованій жировій хворобі печінки основними сайтами, у яких спостерігається аберантне метилювання, є H3K4, H3K9, H3K27, H4R3, H3R8, H2AR3. Ідентифіковано кілька генів гістонових метилтрансфераз та гістонових деметилаз, які диференціально експресуються у хворих з метаболічно асоційованою жировою хворобою печінки та здорових людей. Автори наголошують, що ферменти, які беруть участь у процесах метилювання та деметилювання гістонів, є мішенями майбутніх лікарських засобів, що, безумовно, поліпшить майбутню ефективність медикаментозної терапії у хворих на метаболічно асоційовану жирову хворобу печінки.
Based on the analysis of literary sources of databases Pubmed, MEDLINE, The Cochrane Library, Embase, the authors highlight the essential role of epigenetic modulations in the pathogenesis of metabolic dysfunction-associated fatty liver disease. They give general provisions regarding post-translational histone modifications, which are associated with the development of metabolic dysfunction-associated fatty liver disease, namely, the features of their methylation, post-translational modifications of methylation of lysine residues (sites H3K4, H3K27, H3K36, H3K79, H4K20, H3K23, H3K63 and H4K12), arginine residues (sites H2AR3, H4R3, H3R2, H3R8 and H3R26). Histone methylation and demethylation, mediating the expression of key genes involved in carbohydrate and lipid metabolism, determine both the occurrence and development of steatosis, inflammation and fibrosis of the liver in patients with metabolic dysfunction-associated fatty liver disease. Methylated markers H3K9, H3K27, H4K20 of histones are associated with packed heterochromatin and repression of gene transcription, while methylated markers H3K4, H3K36, H3K79 of histones are associated with activation of gene transcription. Methylation of lysine or arginine residues of histones is carried out by methyltransferases that use S-adenosylmethionine as a donor molecule. The methylated site of an arginine residue can be represented by a monomethylated, asymmetrically demethylated, or symmetrically demethylated marker, whereas demethylation of a lysine marker is mediated by demethylases. The authors provide data that in metabolic dysfunction-associated fatty liver disease, the main sites in which aberrant methylation is observed are H3K4, H3K9, H3K27, H4R3, H3R8, and H2AR3. Several genes of histone methyltransferases and histone demethylases have been identified, which are differentially expressed in patients with metabolic dysfunction-associated fatty liver disease and healthy people. The authors emphasize that the enzymes involved in the histone methylation and demethylation are targets of future drugs, which will certainly improve the effectiveness of drug therapy in patients with metabolic dysfunction-associated fatty liver disease.
ожиріння; метаболічно асоційована жирова хвороба печінки; метилювання гістонів
children; obesity; metabolic dysfunction-associated fatty liver disease; histone methylation
Для ознакомления с полным содержанием статьи необходимо оформить подписку на журнал.
- Абатуров О.Є., Нікуліна А.О. Метаболічно асоційована жирова хвороба/метаболічно асоційована стеатотична хвороба печінки: загальні положення. Здоров’я дитини. 2024;2 (19):68-77. doi: 10.22141/2224-0551.19.2.2024.1683.
- Абатуров О.Є., Нікуліна А.О. Генетична схильність до метаболічно асоційованої жирової хвороби печінки. Здоров’я дитини. 2024;3(19):50-59. doi: 10.22141/2224-0551.19.3.2024.1696.
- Liu Y., Chen M. Histone Methylation Regulation as a Potential Target for Non-alcoholic Fatty Liver Disease. Curr Protein Pept Sci. 2023;24(6):465-476. doi: 10.2174/1389203724666230526155643.
- Dong F., Jiang S., Li J., Wang Y., Zhu L., Huang Y., et al. The histone demethylase KDM4D promotes hepatic fibrogenesis by modulating Toll-like receptor 4 signaling pathway. EBioMedicine. 2019 Jan;39:472-483. doi: 10.1016/j.ebiom.2018.11.055.
- Shi Y., Qi W. Histone Modifications in NAFLD: Mechanisms and Potential Therapy. Int J Mol Sci. 2023 Sep 27;24(19):14653. doi: 10.3390/ijms241914653.
- Kubo N., Chen P.B., Hu R., Ye Z., Sasaki H., Ren B. H3K4me1 facilitates promoter-enhancer interactions and gene activation during embryonic stem cell differentiation. Mol Cell. 2024 May 2;84(9):1742-1752.e5. doi: 10.1016/j.molcel.2024.02.030.
- Liu T., Li R., Sun L., Xu Z., Wang S., Zhou J., et al. Menin orchestrates hepatic glucose and fatty acid uptake via deploying the cellular translocation of SIRT1 and PPARγ. Cell Biosci. 2023 Sep 22;13(1):175. doi: 10.1186/s13578-023-01119-y.
- Kim D.H., Kim J., Kwon J.S., Sandhu J., Tontonoz P., Lee S.K., et al. Critical Roles of the Histone Methyltransferase MLL4/KMT2D in Murine Hepatic Steatosis Directed by ABL1 and PPARγ2. Cell Rep. 2016 Nov 1;17(6):1671-1682. doi: 10.1016/j.celrep.2016.10.023.
- Kim J., Lee B., Kim D.H., Yeon J.G., Lee J., Park Y., et al. UBE3A Suppresses Overnutrition-Induced Expression of the Steatosis Target Genes of MLL4 by Degrading MLL4. Hepatology. 2019 Mar;69(3):1122-1134. doi: 10.1002/hep.30284.
- Rodríguez-Sanabria J.S., Escutia-Gutiérrez R., Rosas-Campos R., Armendáriz-Borunda J.S., Sandoval-Rodríguez A. An Update in Epigenetics in Metabolic-Associated Fatty Liver Disease. Front Med (Lausanne). 2022 Jan 11;8:770504. doi: 10.3389/fmed.2021.770504.
- Tian W., Fan Z., Li J., Hao C., Li M., Xu H., et al. Myocardin-related transcription factor A (MRTF-A) plays an essential role in hepatic stellate cell activation by epigenetically modulating TGF-β signaling. Int J Biochem Cell Biol. 2016 Feb;71:35-43. doi: 10.1016/j.biocel.2015.12.005.
- Gregory G.D., Vakoc C.R., Rozovskaia T., Zheng X., Patel S., Nakamura T., et al. Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol Cell Biol. 2007 Dec;27(24):8466-79. doi: 10.1128/MCB.00993-07.
- Perugorria M.J., Wilson C.L., Zeybel M., Walsh M., Amin S., Robinson S., et al. Histone methyltransferase ASH1 orchestrates fibrogenic gene transcription during myofibroblast transdifferentiation. Hepatology. 2012 Sep;56(3):1129-39. doi: 10.1002/hep.25754.
- Younesi F.S., Miller A.E., Barker T.H., Rossi F.M.V., Hinz B. Fibroblast and myofibroblast activation in normal tissue repair and fibrosis. Nat Rev Mol Cell Biol. 2024 Apr 8. doi: 10.1038/s41580-024-00716-0.
- Garbuzenko D.V. Mechanisms of Epigenetic Regulation in the Fibrogenic Activation of Hepatic Stellate Cells in Non-alcoholic Fatty Liver Disease. Gene Expression 2024;23(1):31-43. doi: 10.14218/GE.2023.00090.
- Zhang L.Y., Wang C.Y., Xu Q., Mu Z.Q., Lin X., Li L.Y., et al. Removal of epigenetic repressive mark on inflammatory genes in fat liver. J Gastroenterol Hepatol. 2023 Aug;38(8):1426-1437. doi: 10.1111/jgh.16252.
- Grewal S.I.S. The molecular basis of heterochromatin assembly and epigenetic inheritance. Mol Cell. 2023 Jun 1;83(11):1767-1785. doi: 10.1016/j.molcel.2023.04.020.
- Jurkowska R.Z., Jeltsch A. Enzymology of Mammalian DNA Methyltransferases. Adv Exp Med Biol. 2022;1389:69-110. doi: 10.1007/978-3-031-11454-0_4.
- Tajima S., Suetake I., Takeshita K., Nakagawa A., Kimura H., Song J. Domain Structure of the Dnmt1, Dnmt3a, and Dnmt3b DNA Methyltransferases. Adv Exp Med Biol. 2022;1389:45-68. doi: 10.1007/978-3-031-11454-0_3.
- Yearim A., Gelfman S., Shayevitch R., Melcer S., Glaich O., Mallm J.P., et al. HP1 is involved in regulating the global impact of DNA methylation on alternative splicing. Cell Rep. 2015 Feb 24;10(7):1122-34. doi: 10.1016/j.celrep.2015.01.038.
- Nakao M., Sato Y., Aizawa A., Kimura H. Mode of SUV420H2 heterochromatin localization through multiple HP1 binding motifs in the heterochromatic targeting module. Genes Cells. 2024 May;29(5):361-379. doi: 10.1111/gtc.13109.
- Jun H.J., Kim J., Hoang M.H., Lee S.J. Hepatic lipid accumulation alters global histone h3 lysine 9 and 4 trimethylation in the peroxisome proliferator-activated receptor alpha network. PLoS One. 2012;7(9):e44345. doi: 10.1371/journal.pone.0044345.
- Yoo J., Jeon Y.H., Cho H.Y., Lee S.W., Kim G.W., Lee D.H., et al. Advances in Histone Demethylase KDM3A as a Cancer Therapeutic Target. Cancers (Basel). 2020 Apr 28;12(5):1098. doi: 10.3390/cancers12051098.
- Theys C., Lauwers D., Perez-Novo C., Vanden Berghe W. PPARα in the Epigenetic Driver Seat of NAFLD: New Therapeutic Opportunities for Epigenetic Drugs? Biomedicines. 2022 Nov 25;10(12):3041. doi: 10.3390/biomedicines10123041.
- Fan L., Sudeep K., Qi J. Histone Demethylase KDM3 (JMJD1) in Transcriptional Regulation and Cancer Progression. Adv Exp Med Biol. 2023;1433:69-86. doi: 10.1007/978-3-031-38176-8_4.
- Qiu Y.Y., Zhang J., Zeng F.Y., Zhu Y.Z. Roles of the peroxisome proliferator-activated receptors (PPARs) in the pathogenesis of nonalcoholic fatty liver disease (NAFLD). Pharmacol Res. 2023 Jun;192:106786. doi: 10.1016/j.phrs.2023.106786.
- Patsouris D., Reddy J.K., Müller M., Kersten S. Peroxisome proliferator-activated receptor alpha mediates the effects of high-fat diet on hepatic gene expression. Endocrinology. 2006 Mar;147(3):1508-16. doi: 10.1210/en.2005-1132.
- Todisco S., Santarsiero A., Convertini P., De Stefano G., Gi–lio M., Iacobazzi V., et al. PPAR Alpha as a Metabolic Modulator of the Liver: Role in the Pathogenesis of Nonalcoholic Steatohepatitis (NASH). Biology (Basel). 2022 May 23;11(5):792. doi: 10.3390/biology11050792.
- Bougarne N., Weyers B., Desmet J., Deckers J., Ray D.W., Staels B., et al. Molecular Actions of PPARα in Lipid Metabolism and Inflammation. Endocr Rev. 2018 Oct 1;39(5):760-802. doi: 10.1210/er.2018-00064.
- Gong L., Wei F., Gonzalez F.J., Li G. Hepatic fibrosis: Targe–ting peroxisome proliferator-activated receptor alpha from mechanism to medicines. Hepatology. 2023 Nov 1;78(5):1625-1653. doi: 10.1097/HEP.0000000000000182.
- Lin Y., Wang Y., Li P.F. PPARα: An emerging target of metabolic syndrome, neurodegenerative and cardiovascular diseases. Front Endocrinol (Lausanne). 2022 Dec 16;13:1074911. doi: 10.3389/fendo.2022.1074911.
- Lefebvre P., Lalloyer F., Baugé E., Pawlak M., Gheeraert C., Dehondt H., et al. Interspecies NASH disease activity whole-genome profiling identifies a fibrogenic role of PPARα-regulated dermatopontin. JCI Insight. 2017 Jul 6;2(13):e92264. doi: 10.1172/jci.insight.92264.
- Unamuno X., Gómez-Ambrosi J., Ramírez B., Rodríguez A., Becerril S., Valentí V., et al. Dermatopontin, A Novel Adipokine Promoting Adipose Tissue Extracellular Matrix Remodelling and Inflammation in Obesity. J Clin Med. 2020 Apr 9;9(4):1069. doi: 10.3390/jcm9041069.
- Catalán V., Domench P., Gómez-Ambrosi J., Ramírez B., Becerril S., Mentxaka A., et al. Dermatopontin Influences the Development of Obesity-Associated Colon Cancer by Changes in the Expression of Extracellular Matrix Proteins. Int J Mol Sci. 2022 Aug 17;23(16):9222. doi: 10.3390/ijms23169222.
- Viscarra J.A., Wang Y., Nguyen H.P., Choi Y.G., Sul H.S. Histone demethylase JMJD1C is phosphorylated by mTOR to activate de novo lipogenesis. Nat Commun. 2020 Feb 7;11(1):796. doi: 10.1038/s41467-020-14617-1.
- Kim J.H., Jung D.Y., Kim H.R., Jung M.H. Histone H3K9 Demethylase JMJD2B Plays a Role in LXRα-Dependent Lipogenesis. Int J Mol Sci. 2020 Nov 5;21(21):8313. doi: 10.3390/ijms21218313.
- Kim J.H., Jung D.Y., Nagappan A., Jung M.H. Histone H3K9 demethylase JMJD2B induces hepatic steatosis through upregulation of PPARγ2. Sci Rep. 2018 Sep 13;8(1):13734. doi: 10.1038/s41598-018-31953-x.
- Skat-Rordam J., Hojland Ipsen D., Lykkesfeldt J., Tveden-Nyborg P. A role of peroxisome proliferator-activated receptor γ in non-alcoholic fatty liver disease. Basic Clin Pharmacol Toxicol. 2019 May;124(5):528-537. doi: 10.1111/bcpt.13190.
- Lee S.M., Muratalla J., Sierra-Cruz M., Cordoba-Chacon J. Role of hepatic peroxisome proliferator-activated receptor γ in non-alcoholic fatty liver disease. J Endocrinol. 2023 Mar 10;257(1):e220155. doi: 10.1530/JOE-22-0155.
- Zaiou M. Peroxisome Proliferator-Activated Receptor-γ as a Target and Regulator of Epigenetic Mechanisms in Nonalcoholic Fatty Liver Disease. Cells. 2023 Apr 21;12(8):1205. doi: 10.3390/cells12081205.
- Chen Q., Peng K., Mo P., Yu C. Histone Demethylase JMJD2D: A Novel Player in Colorectal and Hepatocellular Cancers. Cancers (Basel). 2022 Jun 8;14(12):2841. doi: 10.3390/cancers14122841.
- Li M., Deng Y., Zhuo M., Zhou H., Kong X., Xia X., et al. Demethylase-independent function of JMJD2D as a novel antagonist of p53 to promote Liver Cancer initiation and progression. Theranostics. 2020 Jul 11;10(19):8863-8879. doi: 10.7150/thno.45581.
- Jiang Y., Liu L., Yang Z.Q. KDM4 Demethylases: Structure, Function, and Inhibitors. Adv Exp Med Biol. 2023;1433:87-111. doi: 10.1007/978-3-031-38176-8_5.
- Bricambert J., Alves-Guerra M.C., Esteves P., Prip-Buus C., Bertrand-Michel J., Guillou H., et al. The histone demethylase Phf2 acts as a molecular checkpoint to prevent NAFLD progression during obesity. Nat Commun. 2018 May 29;9(1):2092. doi: 10.1038/s41467-018-04361-y.
- Iizuka K., Bruick R.K., Liang G., Horton J.D., Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci USA. 2004 May 11;101(19):7281-6. doi: 10.1073/pnas.0401516101.
- Ortega-Prieto P., Postic C. Carbohydrate Sensing Through the Transcription Factor ChREBP. Front Genet. 2019 Jun 4;10:472. doi: 10.3389/fgene.2019.00472.
- Iizuka K., Takao K., Yabe D. ChREBP-Mediated Regulation of Lipid Metabolism: Involvement of the Gut Microbiota, Liver, and Adipose Tissue. Front Endocrinol (Lausanne). 2020 Dec 3;11:587189. doi: 10.3389/fendo.2020.587189.
- Benichou E., Seffou B., Topçu S., Renoult O., Lenoir V., Planchais J., et al. The transcription factor ChREBP Orchestrates liver carcinogenesis by coordinating the PI3K/AKT signaling and cancer metabolism. Nat Commun. 2024 Feb 29;15(1):1879. doi: 10.1038/s41467-024-45548-w.
- Régnier M., Carbinatti T., Parlati L., Benhamed F., Postic C. The role of ChREBP in carbohydrate sensing and NAFLD development. Nat Rev Endocrinol. 2023 Jun;19(6):336-349. doi: 10.1038/s41574-023-00809-4.
- Perillo B., Tramontano A., Pezone A., Migliaccio A. LSD1: more than demethylation of histone lysine residues. Exp Mol Med. 2020 Dec;52(12):1936-1947. doi: 10.1038/s12276-020-00542-2.
- Ma T., Li A., Guo Y., Li S., Li M., Feng S., et al. KDM1A/LSD1 as a promising target in various diseases treatment by regulating autophagy network. Biomed Pharmacother. 2022 Apr;148:112762. doi: 10.1016/j.biopha.2022.112762.
- Agarwal S., Bonefas K.M., Garay P.M., Brookes E., Murata-Nakamura Y., Porter R.S., et al. KDM1A maintains genome-wide homeostasis of transcriptional enhancers. Genome Res. 2021 Feb;31(2):186-197. doi: 10.1101/gr.234559.118.
- Yang Z., Zhang S., Liu X., Shu R., Shi W., Qu W., et al. Histone demethylase KDM1A promotes hepatic steatosis and inflammation by increasing chromatin accessibility in NAFLD. J Lipid Res. 2024 Mar;65(3):100513. doi: 10.1016/j.jlr.2024.100513.
- Raas M.W.D., Zijlmans D.W., Vermeulen M., Marks H. There is another: H3K27me3-mediated genomic imprinting. Trends Genet. 2022 Jan;38(1):82-96. doi: 10.1016/j.tig.2021.06.017.
- Van der Meulen J., Speleman F., Van Vlierberghe P. The H3K27me3 demethylase UTX in normal development and disease. Epigenetics. 2014 May;9(5):658-68. doi: 10.4161/epi.28298.
- Kim T.K., Shiekhattar R. Architectural and Functional Commonalities between Enhancers and Promoters. Cell. 2015 Aug 27;162(5):948-59. doi: 10.1016/j.cell.2015.08.008.
- Basta M.D., Petruk S., Mazo A., Walker J.L. Fibrosis-the tale of H3K27 histone methyltransferases and demethylases. Front Cell Dev Biol. 2023 Jul 5;11:1193344. doi: 10.3389/fcell.2023.1193344.
- Zhao F., Ke J., Pan W., Pan H., Shen M. Synergistic effects of ISL1 and KDM6B on non-alcoholic fatty liver disease through the regulation of SNAI1. Mol Med. 2022 Jan 31;28(1):12. doi: 10.1186/s10020-021-00428-7.
- Martin-Mateos R., De Assuncao T.M., Arab J.P., Jalan-Sakrikar N., Yaqoob U., Greuter T., et al. Enhancer of Zeste Homologue 2 Inhibition Attenuates TGF-β Dependent Hepatic Stellate Cell Activation and Liver Fibrosis. Cell Mol Gastroenterol Hepatol. 2019;7(1):197-209. doi: 10.1016/j.jcmgh.2018.09.005.
- Byun S., Seok S., Kim Y.C., Zhang Y., Yau P., Iwamori N., et al. Fasting-induced FGF21 signaling activates hepatic autophagy and lipid degradation via JMJD3 histone demethylase. Nat Commun. 2020 Feb 10;11(1):807. doi: 10.1038/s41467-020-14384-z.
- Liu Y., Chen M. Histone Demethylation Profiles in Nonalcoholic Fatty Liver Disease and Prognostic Values in Hepatocellular Carcinoma: A Bioinformatic Analysis. Curr Issues Mol Biol. 2023 Apr 20;45(4):3640-3657. doi: 10.3390/cimb45040237.
- Yang Y., Chen X.X., Li W.X., Wu X.Q., Huang C., Xie J., Zhao Y.X., et al. EZH2-mediated repression of Dkk1 promotes hepatic stellate cell activation and hepatic fibrosis. J Cell Mol Med. 2017 Oct;21(10):2317-2328. doi: 10.1111/jcmm.13153.
- Panebianco C., Oben J.A., Vinciguerra M., Pazienza V. Senescence in hepatic stellate cells as a mechanism of liver fibrosis reversal: a putative synergy between retinoic acid and PPARgamma signalings. Clin Exp Med. 2017 Aug;17(3):269-280. doi: 10.1007/s10238-016-0438-x.
- Sinha R.A. Targeting nuclear receptors for NASH/MASH: From bench to bedside. Liver Res. 2024 Mar;8(1):34-45. doi: 10.1016/j.livres.2024.03.002.
- Thibonnier M., Esau C. Metabolic Benefits of MicroRNA-22 Inhibition. Nucleic Acid Ther. 2020 Apr;30(2):104-116. doi: 10.1089/nat.2019.0820.
- Seok S., Kim Y.C., Byun S., Choi S., Xiao Z., Iwamori N., et al. Fasting-induced JMJD3 histone demethylase epigenetically activates mitochondrial fatty acid β-oxidation. J Clin Invest. 2018 Jul 2;128(7):3144-3159. doi: 10.1172/JCI97736.
- Kosgei V.J., Coelho D., Guéant-Rodriguez R.M., Guéant J.L. Sirt1-PPARS Cross-Talk in Complex Metabolic Diseases and Inherited Disorders of the One Carbon Metabolism. Cells. 2020 Aug 11;9(8):1882. doi: 10.3390/cells9081882.
- Grindheim J.M., Nicetto D., Donahue G., Zaret K.S. Polycomb Repressive Complex 2 Proteins EZH1 and EZH2 Regulate Timing of Postnatal Hepatocyte Maturation and Fibrosis by Repressing Genes with Euchromatic Promoters in Mice. Gastroenterology. 2019 May;156(6):1834-1848. doi: 10.1053/j.gastro.2019.01.041.
- Basta M.D., Petruk S., Mazo A., Walker JL. Fibrosis-the tale of H3K27 histone methyltransferases and demethylases. Front Cell Dev Biol. 2023 Jul 5;11:1193344. doi: 10.3389/fcell.2023.1193344.
- Peng W., Cheng S., Bao Z., Wang Y., Zhou W., Wang J., et al. Advances in the research of nanodrug delivery system for targeted treatment of liver fibrosis. Biomed Pharmacother. 2021 May;137:111342. doi: 10.1016/j.biopha.2021.111342.
- Argemi J., Bataller R. Identifying New Epigenetic Drivers of Liver Fibrosis. Cell Mol Gastroenterol Hepatol. 2019;7(1):237-238. doi: 10.1016/j.jcmgh.2018.09.015.
- Chaturvedi S.S., Ramanan R., Waheed S.O., Karabencheva-Christova T.G., Christov C.Z. Structure-function relationships in KDM7 histone demethylases. Adv Protein Chem Struct Biol. 2019;117:113-125. doi: 10.1016/bs.apcsb.2019.08.005.
- Li C.Y., Liu Y.J., Tao F., Chen R.Y., Shi J.J., Lu J.F., et al. Lysine-specific demethylase 7A (KDM7A): A potential target for disease therapy. Biochem Pharmacol. 2023 Oct;216:115799. doi: 10.1016/j.bcp.2023.115799.
- Wang Y., Hong Q., Xia Y., Zhang Z., Wen B. The Lysine Demethylase KDM7A Regulates Immediate Early Genes in Neurons. Adv Sci (Weinh). 2023 Oct;10(28):e2301367. doi: 10.1002/advs.202301367.
- Kim J.H., Nagappan A., Jung D.Y., Suh N., Jung M.H. Histone Demethylase KDM7A Contributes to the Development of Hepatic Steatosis by Targeting Diacylglycerol Acyltransferase 2. Int J Mol Sci. 2021 Oct 14;22(20):11085. doi: 10.3390/ijms222011085.
- Zaiou M., Amrani R., Rihn B., Hajri T. Dietary Patterns Influence Target Gene Expression through Emerging Epigenetic Mechanisms in Nonalcoholic Fatty Liver Disease. Biomedicines. 2021 Sep 18;9(9):1256. doi: 10.3390/biomedicines9091256.
- Amin N.B., Carvajal-Gonzalez S., Purkal J., Zhu T., Crowley C., Perez S., et al. Targeting diacylglycerol acyltransferase 2 for the treatment of nonalcoholic steatohepatitis. Sci Transl Med. 2019 Nov 27;11(520):eaav9701. doi: 10.1126/scitranslmed.aav9701.
- Kim H., Ronai Z.A. PRMT5 function and targeting in cancer. Cell Stress. 2020 Jul 13;4(8):199-215. doi: 10.15698/cst2020.08.228.
- Motolani A., Martin M., Sun M., Lu T. The Structure and Functions of PRMT5 in Human Diseases. Life (Basel). 2021 Oct 12;11(10):1074. doi: 10.3390/life11101074.
- Zheng J., Li B., Wu Y., Wu X., Wang Y. Targeting Arginine Methyltransferase PRMT5 for Cancer Therapy: Updated Progress and Novel Strategies. J Med Chem. 2023 Jul 13;66(13):8407-8427. doi: 10.1021/acs.jmedchem.3c00250.
- Shang S., Li X., Azzo A., Truong T., Dozmorov M., Lyons C., et al. MBD2a-NuRD binds to the methylated γ-globin gene promoter and uniquely forms a complex required for silencing of HbF expression. Proc Natl Acad Sci USA. 2023 Jun 20;120(25):e2302254120. doi: 10.1073/pnas.2302254120.
- Huang L., Liu J., Zhang X.O., Sibley K., Najjar S.M., Lee M.M., et al. Inhibition of protein arginine methyltransferase 5 enhances hepatic mitochondrial biogenesis. J Biol Chem. 2018 Jul 13;293(28):10884-10894. doi: 10.1074/jbc.RA118.002377.
- Zhang Y., Verwilligen R.A.F., Van Eck M., Hoekstra M. PRMT5 inhibition induces pro-inflammatory macrophage polarization and increased hepatic triglyceride levels without affecting atherosclerosis in mice. J Cell Mol Med. 2023 Apr;27(8):1056-1068. doi: 10.1111/jcmm.17676.